Ribosome

The ribosome is the molecular machine inside the cell that makes proteins from amino acids in the process called translation . It binds to a messenger ribonucleic acid (mRNA) and reads the information contained in the nucleotide sequence of the mRNA. Transfer RNAs (tRNAs) containing amino acids enter the ribosome in a special pocket, or binding site, called the acceptor site (A site). Once correctly bound, the ribosome can add the amino acid on the tRNA to the growing protein chain.
Structure
The ribosome is made up of two parts, called subunits. The larger of the two subunits is where the amino acids get added to the growing protein chain. The small subunit is where the mRNA binds and is decoded. Each of the subunits is made up of both protein and ribonucleic acid (RNA) components.
The small ribosomal subunit is made up of one ribosomal RNA (rRNA) and approximately twenty-one proteins in prokaryotes (bacteria) and approximately thirty-three proteins in eukaryotes (mammals). In prokaryotes, the large ribosomal subunit contains two rRNAs—one large one and one small one—and approximately thirty-one proteins. In eukaryotes, the large subunit is composed of three rRNAs—one large one and two different small ones—and approximately forty-nine proteins. In eukaryotic cells, ribosomal subunits are synthesized in the nucleolus and then exported to the cytoplasm before use.
The rRNAs have many regions of self-complementarity, that is, regions within the rRNA that can form base pairs with other regions of the same rRNA, linking them together. This self-complementarity produces highly
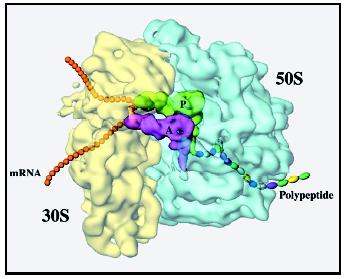
Researchers have worked for many years to try to determine what the ribosome's structure is at the atomic level. How are all the atoms that make up the ribosome arranged in three-dimensional space? On a gross level, the ribosome looks something like an oyster with one of its shells somewhat smaller than the other. The two subunits are joined to each other by interactions between the rRNAs in one subunit and proteins in the other subunit. There may also be interactions between an RNA on one subunit and an RNA on the other subunit and between proteins on the two subunits.
RNA Movements
tRNAs move through the ribosome during the course of protein synthesis. A tunnel runs through the ribosome, right at the interface between the two subunits, and the tRNAs enter one side of this tunnel and are propelled along it during each step of protein synthesis. The three tRNA binding sites of the ribosome—A (acceptor), P (peptidyl), and E (exit)—appear to be intermediate spots in this tunnel. The mRNA binds to a groove at the bottom of the tRNA tunnel. After each amino acid is added to the growing protein, the tRNAs must be moved from one site to the next, and the mRNA must also be moved over one codon (three bases) so that the next amino acid coded for by the mRNA can be added to the protein.
These movements of the tRNAs and mRNA are made possible by a protein factor, called EF-G in prokaryotes or EF-2 in eukaryotes, which binds to the ribosome and uses the energy stored in the triphosphate group of guanosine triphosphate (GTP) to help propel the tRNAs and mRNA along. It also appears that parts of the ribosome move as the tRNAs and mRNA move. In fact, it is possible that EF-G produces movement of these parts of the ribosome and that these movements in turn produce movement of the tRNAs and mRNAs. Certain antibiotics (drugs that kill bacteria) are known to work by preventing some of the movements of bacterial ribosomes, thus stopping protein synthesis.
Intriguingly, there are certain mutations of the ribosome (changes to the structure of the rRNA or proteins) that affect its movements during translation and appear to cause a decrease in the accuracy of protein synthesis (for example, the wrong amino acids get put into the protein with increased frequency). Thus, the movements themselves may be directly tied to the mechanism by which the ribosome makes sure that the correct amino acid is being added to the protein at each point along the mRNA.
The growing protein chain exits the ribosome through a second tunnel, this one at the top of the large subunit. When protein synthesis ends, the binding of proteins called release factors is thought to induce the ribosome to release the finished protein into the cytoplasm. Exactly how the ribosome does this is unclear.
For many years it was thought that the rRNAs in the ribosome served merely as a scaffold on which to hang the ribosomal proteins. It was proposed that the proteins did all of the important work in the ribosome, such as catalyzing the formation of peptide bonds and moving the tRNAs and mRNA along during protein synthesis. However, it is now clear that the rRNAs play an active role in protein synthesis and are not merely the frame on which the ribosome is built. As more detailed information about the three-dimensional structure of the ribosome becomes available and as researchers do more experiments to probe the inner workings of this fascinating machine, we will have a better understanding of what the rRNAs do and how they do it.
SEE ALSO Nucleolus ; Protein Synthesis ; RNA ; Transfer RNA
Jon Lorsch
Bibliography
Cate, Jamie H., et al. "X-ray Crystal Structures of 70S Ribosome Functional Complexes." Science 285 (1999): 2095–2104.
Frank, Joachim. "How the Ribosome Works." American Scientist 86, no. 5 (1998): 428–439.
Hill, Walter E., et al., eds. The Ribosome: Structure, Function & Evolution . Washington, DC: American Society for Microbiology Press, 1990.
Lewin, Benjamin. Genes VI. Oxford: Oxford University Press, 1997.
Puglisi J. D., S. C. Blanchard, and R. Green. "Reviews—Approaching Translation at Atomic Resolution." Nature Structural Biology 7, no. 10 (2000): 855–861.
The poison ricin, from castor bean seeds, cleaves part of the RNA in the large subunit.
Comment about this article, ask questions, or add new information about this topic: