Genomics
Genomics is the study of the genome , or deoxyribonucleic acid (DNA), of an organism and associated technologies. Genomics evolved from a series
Before the development of genomics, DNA studies in humans were mostly relegated to observing highly condensed chromosomes under a microscope. Many human studies were done in medical centers where families with visible chromosomal mutations came for evaluation. While it was possible in a few cases to determine on which chromosome a gene was located or whether two genes were located on the same chromosome, without molecular techniques these were difficult tasks at best.
Tools of Genomics
Genomics developed from advances in recombinant DNA technology, while in turn, developed from earlier progress in biochemistry and genetics. Recombinant DNA technology is the set of tools that make it possible for researchers to study and manipulate DNA, ribonucleic acid (RNA), and protein from any source, both outside of the cells ( in vitro ) and inside of the cells ( in vivo ) of the well-studied model organisms.
Relatively few techniques are used to study DNA. The basic methods that underlie genomic technologies include DNA sequencing, polymerase chain reaction (PCR), electrophoresis , cloning, and hybridization. The fact that all DNA molecules can be manipulated using a few basic techniques is a major advantage of working with DNA. In contrast, proteins are much more difficult to manipulate, and each must be approached individually.
DNA sequencing determines the order of bases in a segment of DNA, a gene, a chromosome, or an entire genome. PCR can increase the number of copies (even a millionfold) of a single gene or fragment of DNA in vitro within hours. Electrophoresis separates DNA by size in the presence of an electrical field. This is a simple technique used to follow changes in DNA size through different recombinant manipulations.
Cloning
The process of isolating a piece of DNA for recombinant DNA studies is called cloning. Cloning increases the number of copies of a single gene or fragment of DNA in vivo . Large amounts of DNA are needed for sequencing and manipulation experiments. The purified, unknown DNA is combined with another well-characterized piece of DNA called a cloning vector . The vector DNA has the DNA sequences needed to form an artificial minichromosome. A cell that contains the cloned DNA is called a clone. The clone is used to produce more copies of the DNA of interest and to produce protein encoded by the DNA. In some experiments, a modified DNA segment is returned to the original organism for further studies.
Cloning can be used to isolate and scrutinize a small part of the genome, such as a gene. For example, consider the cloning of an oncogene , which is a gene that, when overexpressed, causes cancer. DNA fragments from human cancer cells were introduced into the cells of a normal mouse. Some of the cells received a piece of human DNA that caused them to develop into cancerous cells. Recombinant DNA techniques were used to identify which piece of human DNA was responsible for converting the normal cells to cancerous cells. DNA methods enabled researchers to isolate the specific human gene that causes the cancer.
Hybridization
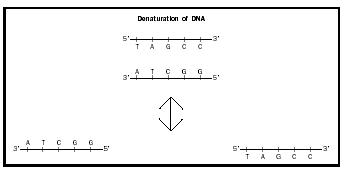
Hybridization, which is also known as renaturation or annealing, is the coming together of two complementary , single strands of DNA to form double-stranded DNA. Denaturation is the reverse process, which separates double-stranded DNA into two single strands (see Fig. 1). Double-stranded DNA is denatured when it is incubated at a high temperature. In hybridization experiments, single-stranded DNA of unknown sequence (test DNA) is hybridized to single-stranded DNA of known sequence (probe DNA). Hybridization takes place only when the test DNA contains a complementary DNA sequence.
Hybridization experiments can be used to sequence the test DNA a "word" at a time. A word is equal to the length of the probe sequence, which is usually greater than eight bases. This technique makes it possible to obtain information more rapidly than from ordinary sequencing methods.
DNA hybridization experiments are also used to study the expression of thousands of genes at the same time. In expression studies, messenger RNA (mRNA) from a cell is hybridized to an array (display) of complementary single-stranded DNA probe sequences unique for each test mRNA. Test mRNA hybridizes to DNA probes in the same manner as test DNA. The amount of test mRNA hybridized to each probe is measured to determine the amount of complementary mRNA present. Since the hybridization is done to the entire array simultaneously, information about all test mRNA sequences is obtained in a single experiment. The DNA probes arrayed on a microchip are recorded and the hybridization results are analyzed automatically using computers.
DNA array experiments identify and analyze mRNAs solely on the basis of their sequence. No information is needed about the proteins encoded
Finding Disease Genes
Positional cloning experiments isolate genes responsible for a specific genetic disease, such as cystic fibrosis, by first identifying their location (position) in the genome (which chromosome they are on). Genetic mapping techniques locate a gene or a DNA sequence among the chromosomes by identifying which regions of the genome are inherited in the same manner as the trait of interest, such as a disease. Once the chromosomal region is found, molecular and computational methods are used to identify all genes in the region and to pick up "candidate" genes for further testing. Some of the candidate genes are identified by determining whether their encoded proteins fit the trait or disease of interest. For example, the isolation and determination of the DNA sequence of the cystic fibrosis gene made it possible to identify its product, a large protein molecule that regulates the transport of chloride across the cell membrane. Researchers were able to explain the clinical symptoms when they discovered that the protein was nonfunctional.
The power of focusing on genetic causes of disease is due to several factors. First, positional cloning is guaranteed to produce results provided enough money is available to do the experiment and large families inheriting the trait of interest are known. It is very rare in research to be sure of success. Second, the identification of a gene responsible for a disease uncovers pathways and genes that are unknown and may play a role in noninherited forms of the same or a similar disease.
Future Directions
The first stage of genomic research is coming to an end as the entire human DNA sequence is known. An understanding of that blueprint will likely take many more years of research. That understanding will require sequencing many genomes to determine how variations in DNA sequences affect protein and cell function. Further, each gene must be understood in the context of the entire repertoire of thirty-five thousand human genes. Because of advances in genomics, scientists are no longer forced to study single genes out of context. Consequently, experiments and the information gathered in each experiment are becoming more complex. New computational tools are needed to understand the massive amount of information that is now being generated. This has developed into the field of bioinformatics.
The DNA sequences of any two individuals can differ in at least one million places. In medicine, variations in the DNA sequences will be used to develop individualized drug treatments. This new field, known as pharmacogenomics, is in its infancy, but already DNA profiles are being used to subtype different cancers, enabling physicians to prescribe the drugs most likely to be effective for a particular patient in the course of treatment.
Genomic studies are not confined to humans, but are used to learn about all organisms. The lessons that nature will provide from all these studies will have an impact on fields far beyond biology and medicine. The tools and knowledge needed to accomplish these cross-traditional boundaries of scientific disciplines. The solutions to future scientific problems will require an immense amount of collaboration and will need to take advantage of talents and knowledge of a large number of individuals.
SEE ALSO Bioinformatics ; Clone ; DNA Sequencing ; Human Genome Project ; Hybridization ; Model Organisms: Cell Biology and Genetics ; Oncogenes and Cancer Cells ; Polymerase Chain Reaction ; Recombinant DNA
Cassandra L. Smith and Linda G. Tolstoi
Bibliography
Biomedicine Promises for Health. Washington, DC: Pharmaceutical Research and Manufacturers of America (PHRMA), 1998. http://www.phrma.org/publications/publications/brochure/biomed/ .
Casey, Denise. Primer on Molecular Genetics. Washington, DC: U.S. Department of Energy, Office of Energy Research, Office of Health and Environmental Research, 1992.
Hamadeh, Hisham, and Cynthia A. Afshari. "Gene Chips and Functional Genomics." American Scientist 88, no. 6 (2000): 508–515.
Haseltine, William A. "Discovering Genes for New Medicines." Scientific American 276, no. 3 (1997): 92–97.
Lewis Ricki. Human Genetics: Concepts and Applications, 3rd ed. Boston: WCB/Mc-Graw-Hill, 1999.
Tolstoi, Linda, and Cassandra L. Smith. "Human Genome Project and Cystic Fibrosis—a Symbiotic Relationship." Journal of the American Dietetic Association 99, no. 11 (1999): 1421 – 1427.
Vaughan, Douglas. To Know Ourselves. Washington, DC: U.S. Department of Energy and the Human Genome Project, 1996. http://www.ornl.gov/hgmis/ .
Comment about this article, ask questions, or add new information about this topic: